
Introduction
It’s not often that one can witness the birth of a new technological paradigm. One such moment occurred in 1959 at the California Institute of Technology when Nobel Prize-winning physicist Richard Feynman delivered his now seminal talk “There’s Plenty of Room at the Bottom.” 1 1 A video recording of a similar lecture he gave on the topic in the 1980s can be found on Youtube. Expand Footnote Collapse Footnote
He began, not with grand equations or complicated terminology, but with a question: Why can’t we write the entire twenty-four volumes of the Encyclopedia Britannica on the head of a pin? With this seemingly whimsical question, he led the audience down the rabbit hole towards the concept that would come to be called nanotechnology. 2 2 The term 'nano' has its roots in the Greek word 'Nanos', which translates to 'Dwarf'. It describes a fraction that is one billionth of the original unit. Expand Footnote Collapse Footnote Declaring that “the principles of physics, as far as I can see, do not speak against the possibility of maneuvering things atom by atom,” Feynman introduced the possibility of manipulating matter at its most fundamental level, dreaming of tiny machines that could build materials, tools, and products with entirely new properties.
According to the National Nanotechnology Initiative, nanotechnology involves the manipulation of objects from 0.1 nanometers (individual atoms) to 100 nanometers (anything that can’t be seen with a regular optical microscope). To put this into perspective, 1 nanometer is one-billionth of a meter, which is between two to twenty atoms. 3 3 Depending on the atom. Expand Footnote Collapse Footnote
Modern civilization has succeeded at building commercial applications at the larger end of the spectrum. But working at the smaller end, would mean manipulating objects with unprecedented accuracy. Nanotechnology at this tiny scale is called “molecular nanotechnology” or “atomically precise nanotechnology.” To understand the potential, one just needs to remember that atoms are the fundamental building blocks of the material world. Since humans first smashed rocks together to make tools, we have been engineering atoms “in unruly herds” rather than as individuals. 4 4 “Unruly herds” was used by Eric Drexler in Engines of Creation. Expand Footnote Collapse Footnote
Molecular manufacturing, in its most ambitious incarnation, would use programmable tools to position single atoms and create specific chemical bonds in order to build larger structures from the ground up. This would be true precision engineering. 5 5 Precision is extremely important for the progress of technology. For instance, see Simon Winchester’s The Perfectionists. Expand Footnote Collapse Footnote Instead of relying on expensive and scarce resources, molecular manufacturing would take advantage of the abundant nanoscale elements around us. 6 6 A more detailed discussion of the precision, waste, and cost benefits of molecular nanotechnology can be found in Unbounding the Future by Eric Drexler, Christine Peterson and Gayle Pergamit. Expand Footnote Collapse Footnote
With new materials, we could build machines that function in harsh environments, such as outer space. Or we could revolutionize the production of photovoltaics, semiconductors, and other electronics. Our medicine would become far more personalized and powerful. In his lecture, Feynman discusses nanocomputers that would vastly exceed ordinary computers in efficiency and speed. Better computers may, in turn, help us design better nanotechnology, further accelerating the flywheel of progress. Asking what nanotechnology can do with atoms is akin to asking what computers can do with bits—almost every industry would undergo a radical transformation.
According to technology analyst Eli Dourado, the impact of molecular manufacturing on the economy would be hard to exaggerate. Nanotechnology pioneers Josh Storrs Hall and Robert Freitas “each independently and simultaneously estimated that mature nanotechnology could replicate the entire capital stock of the United States – ‘every single building, factory, highway, railroad, bridge, airplane, train, automobile, truck, and ship’ – in a mere week… that would correspond to a US GDP of four quadrillion dollars per year, around 160 times as high as it is today.” 7 7 Dourado continues that “If you take the value of the capital stock at roughly $80 trillion and ignore compounding, that would correspond to a US GDP of four quadrillion dollars per year, around 160 times as high as it is today. Is such a thing possible?” Expand Footnote Collapse Footnote
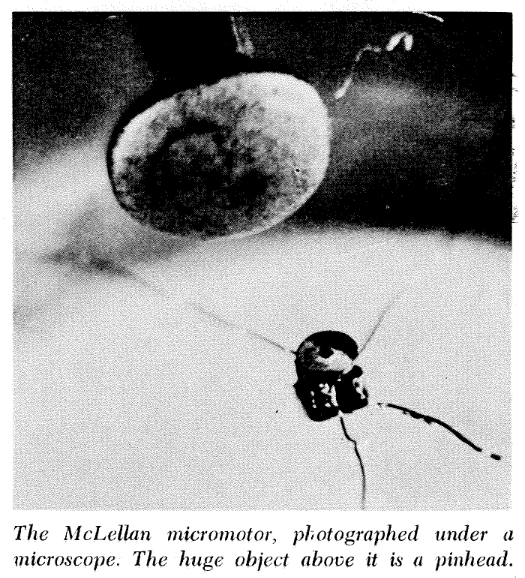
Feynman hoped that this new field was not just a possibility but an inevitability: “in the year 2000, when they look back at this age, they will wonder why it was not until the year 1960 that anybody began seriously to move in this direction." In his lecture, Feynman offered cash prizes for the first teams to produce nanoscale objects, a tiny motor and a tiny book. 8 8 In his lecture, Feynman intended to offered a prize of $1,000 for the first individual to devise a method for reducing a page of text to a size only readable under an electron microscope, and another $1000 prize for constructing a working motor measuring no more than 1/64th of an inch per side. Inspired by Feynman’s desire to incentivize research, Foresight’s annual Feynman Prizes reward useful work that furthers the long-term goal of nanotechnology. Expand Footnote Collapse Footnote Less than a year later, William McLellan produced the motor, and in 1985, Tom Newman produced the book. Since then, there have been significant achievements in nanotechnology. Modern vaccines take advantage of lipid nanoparticles. New materials with remarkable properties, such as carbon nanotubes and graphene, are used in aircraft and next-generation electronic devices. But despite advances, we are still nowhere close to actually building macroscale objects by “maneuvering things atom by atom.” We have failed to realize the full scope of Feynman’s dream.
To understand why, we need to understand the two major pathways that emerged after Feynman. “Protein engineering” involves designing and synthesizing custom proteins that perform specific tasks at the molecular level. Such proteins effectively become the machinery for molecular-scale operations and assemblies. This is the bottom up approach, where tiny systems build their way up to larger products. “Mechanosynthesis” is the direct, positional assembly of atomic structures using specialized tools. This is the top-down approach, where building blocks are arranged atom-by-atom. It now seems that the most hopeful avenue of development lies closer to the middle of these two approaches by creating something akin to a “molecular 3D printer.” In practice, such research has been stunted by technical complexities, a constrained funding landscape, public misunderstanding, and coordination problems endemic to fundamental scientific research. An exploration of these factors shows how contingent they really are—there’s no reason why we can’t achieve molecular manufacturing within our lifetimes.
Paths to Molecular Manufacturing
Protein Engineering
Independently to Feynman’s vision, Eric Drexler released a paper in 1981 detailing a different path towards molecular manufacturing. While Feynman suggested machines that could build smaller and more precise machines recursively all the way down to the nanoscale, Drexler envisioned a bottom-up alternative. After all, he thought, we already have molecular machines in every cell of the human body!
In order to make the proteins necessary for life, all cells rely on molecular machines called ribosomes. They build complex proteins out of building blocks according to specified instructions. In order to get these genetic instructions to the ribosome, the DNA information inside the cell’s nucleus is converted to mRNA. The ribosome then takes the mRNA and, using the free-floating amino acids within the cell, creates the specific chain of amino acids that constitute the needed protein. Finally, the protein folds into a 3D structure that is ready to carry out a variety of intracellular functions. Importantly, before the cell divides, a ribosome will create all the necessary proteins to form another ribosome. The ribosome is a molecular machine that builds other molecular machines. 9 9 Other factors are necessary for ribosome biogenesis, such as the assembly of ribosomal proteins and RNA into subunits within the nucleolus. But the construction of the ribosomal proteins are carried out by the ribosome outside of the cell nucleus. Expand Footnote Collapse Footnote
Ribosomes aren’t the only impressive part of our intracellular machinery; the proteins which ribosomes produce are also essential. Proteins are the fundamental building block of life, performing roles within the cell that are strikingly akin to machinery: motors, actuators, cables, pumps, conveyor belts, and assembly lines. The ability to custom-design proteins could pave the way for the construction of even more complex molecular objects via external instructions. The idea of using proteins as the fundamental units of production for versatile nanoscale machines and assembly lines can be called the protein engineering route to molecular manufacturing.
To engineer a protein, one could design the sequence of amino acids in the chain and encode the instructions as genetic material and hand it to a ribosome. 10 10 Eli Dourado points out that the second process is now employed by mRNA vaccines. Expand Footnote Collapse Footnote There are at least two major stumbling blocks to engineering proteins: the sheer number of possible combinations of amino acids, and the protein folding problem. As Drexler wrote, “an engineer designing a protein that has 1000 amino acids may choose among some 101300 different amino acid sequences.” To put that in perspective, there are roughly 1078 to 1082 atoms in the observable universe! Not only are the possible combinations of amino acids enormous, but it is very difficult to predict how proteins might fold into 3D shapes based on the sequence of their constituent amino acids. In other words, the problem of nanotechnology is not merely a hardware problem but also a software one. The nanoscale world is vast and complex, requiring large amounts of computation in order to be accessible to humans.
Luckily, two recent technological trends could lend a hand: the plummeting cost of computation and the rise of AI artificial intelligence. There have been massive advancements in solving protein folding. In 2020, DeepMind's AlphaFold 2 predicted protein structures with remarkably high accuracy, and we now have an enormous protein database of structural predictions. In addition, the Baker Lab builds up on this effort by leveraging deep learning to produce entire molecular design pipelines, where structures are proposed and matching proteins generated, in order to produce a library of computationally designed protein machines. Molecular machines are Drexler’s “engines of creation,” while AI assistance would be the “engines of design.”
Let’s imagine the design hurdles were overcome: we would now be in a position to build proteins that could reliably fold into stable shapes as necessary, acting as workhorses capable of carrying out diverse tasks. One of their most important functions would be as programmable constructors that build more complex structures, materials, and machines. For that, they need a variety of tools to manipulate and supervise their environment, similar to a traditional factory supervisor observing the machinery and adjusting the production process when necessary.
In 2016, the Nobel Prize in Chemistry was awarded “for the design and synthesis of molecular machines” such as molecular elevators, motors, and even a “nanocar.” However, these machines were assembled in a bespoke manner rather than built up by a more general method. They were not composed of proteins, but synthesized molecules. This makes their use cases limited and their production labor-intensive compared to what protein engineering could produce. Ultimately, we should be interested in the methods of production used to make molecular machines. We need machines that can make machines, not “just” end products.
Even if we were able to design more general molecular tools and machines using protein-engineering, one future problem is that ribosomes operate inside watery cells, which means that they can’t function in high or low temperatures where water evaporates or freezes. In order to work in diverse environments, later generations of more advanced molecular machines would need to be constructed out of inorganic materials. Hopefully we can eventually transition from organic ribosomes to artificial ones that are more durable, precise, and controllable.
Mechanosynthesis
An alternative approach that emerged in parallel to the protein engineering path aims to build complex macro structures by directly manipulating atoms mechanically, often using atomic force microscopy (AFM) or scanning tunneling microscopy (STM). 11 11 To be precise, mechanosynthesis often involves positioning small reactive molecular fragments in order to manipulate single atoms. AFM and STM are used to examine materials at the atomic and molecular levels. While AFM relies on the interaction forces between the tip and the sample surface, STM uses a conductive tip to scan the surface of a sample. Expand Footnote Collapse Footnote In 1989, IBM’s Don Eigler and collaborators used STM to move single atoms and bind them to specific sites on a surface, creating custom patterns, such as spelling the letters “IBM” out of 35 individual atoms. This was one of the first attempts to build at the atomic level, showing that properties of atoms, such as their electric or magnetic charges, could be leveraged to build simple structures. 12 12 By 2002, Eigler’s group had even made patterns of atoms that could create tiny logic gates. Eigler and his team strategically positioned individual carbon monoxide molecules on a flat copper surface. They then instigated a chain reaction by relocating some of these molecules, causing them to collide with other molecules. The final arrangement of these molecules represented the solution to the computation being carried out. Although not economical, Eigler demonstrated that calculations could be performed on a molecular scale. Expand Footnote Collapse Footnote
Continuing Eigler’s work, IBM published A Boy and His Atom in 2013, which holds a world record for the smallest stop-motion film. It portrays a cartoon boy playing with a ball, with each frame individually created by positioning CO molecules on a copper surface and magnifying each atom more than 100 million times.

As entertaining as the resulting imagery may be, is the underlying ability to position individual atoms and molecules useful? It is certainly more precise than conventional chemistry methods, allowing for unprecedented fine-grained control. Using mechanical force to precisely position elements to create more complex assemblages is called “mechanosynthesis.”
Although mechanosynthesis is incredibly precise, such precision comes at the cost of scale—positioning atoms one-by-one is a labor intensive process. In comparison, the trillions of ribosomes in the human body demonstrate their ability to produce proteins at scale. 13 13 Eli Dourado notes that “ribosomes produce proteins much faster than our state-of-the-art lab equipment – human ribosomes can bond about two amino acids per second (and bacterial ribosomes are yet faster), while tabletop reactions happen at a rate of one bond every 2.5 minutes.” Expand Footnote Collapse Footnote Ribosomes can build and verify other molecular machines without relying on continuous supervision. In order for mechanosynthesis to scale, numerous microscopes working in parallel would be required, with each tiny tool somehow being controlled by a macroscopic mechanism.
An early attempt to create automated mechanosynthesis was the nanofactory collaboration. The group intended to use mechanosynthesis to build structures out of diamondoid materials that are strong and durable even at extremely small scales. These specialized “nanofactories” were intended to build macroscale products from the atom up, including other nanofactories. Philip Moriarty, who worked with the nanofactory project, described some particular difficulties: diamond is a particularly hard surface to work with that requires specialized tip and surface preparation. Ultimately, the team switched their materials to silicon and seemingly ceased working on single-atom positioning.
Early mechanosynthesis attempts are comparable to building up a computer processor by hand, transistor by transistor, rather than using modern manufacturing processes. Furthermore, simple 2D assembly that pushes or slides atoms across a surface is a long way from building complex 3D structures which require long sequences of interactions. Direct atomic manipulation is a useful proof of concept, but it would be an extremely slow, fragile, and tedious path for reaching a molecular manufacturing future.
The Molecular 3D Printer
The protein engineering approach is limited by the vast design space and lack of a clear development path from organic to inorganic molecular machines. The mechanosynthesis approach, although more empirically validated, would be hard to scale into a fully automated assembly line that could produce useful products.
This is why nanotechnology enthusiasts have been focusing on an alternative approach to molecular manufacturing: a biomolecular 3D printer operating in a solution that is programmed to guide the bonding of nanoscale building blocks. 14 14 See Eric Drexler’s book Nanosystems, the Foresight roadmap, and a recent conversation with Give Well. Expand Footnote Collapse Footnote Like a conventional 3D printer, it would need an active printer head that can be controlled. For instance, its actuation signal could come from stimuli in the solution that triggers the head to move. The head would print by selectively activating binding sites on chemical building blocks which would bond to one another after being jostled into position by Brownian motion.
If successful, this 3D printer operating in a liquid solution might give way to one which can operate in gaseous, “dry” environments. In liquids, Brownian motion is much more predictable with higher levels of interaction between molecules. Building up to a dry printer may involve intermediate versions that mechanically control the position of materials with only minimal reliance on Brownian motion. However, a dry printer would permit the use of more diverse materials and assembly techniques, such as molecules containing hydrogen, carbon, or oxygen.
In a 2016 workshop hosted by Adam Marblestone and Eric Drexler, they underlined the importance of having a general printer architecture which would outline the various components and technical challenges to be addressed. 15 15 See this interview about the 2016 workshop that inspired Marblestone’s 3D printer prototype. Expand Footnote Collapse Footnote Each of these challenges could then be worked on by multiple labs utilizing different approaches, thereby reducing risk. Marblestone has outlined three key components of a potential molecular printer: a rigid 3D framework for structural support, three axes of programmable motion control, and building blocks which "snap" into defined binding sites on a printed substrate.
The body of the 3D printer could be built from “DNA origami”, taking advantage of the structural properties of the double helix to create tiny, robust frames. DNA origami leverages Watson-Crick base pairing rules to turn single-stranded DNA into precise structures. 16 16 DNA origami is in full swing: One of the earliest approaches, ‘Scaffolded DNA origami’, was developed by Paul Rothemund. It linked multiple DNA helices into complex shapes by using DNA ‘staple strands’ to connect parts of longer ‘scaffold strands’. William Shih further developed this technique to create fully addressable 3D DNA origami structures, where each staple strand can be attached to a specific molecule to position it within a larger structure. Expand Footnote Collapse Footnote Andrew Turberfield and his team have combined three independently controllable DNA origami actuators to create a printer that can read and write to a DNA nanofabric. They started with a moving gantry that runs across two parallel rails. First, they showed that, when signaling DNA molecules are added, the gantry is capable of reversibly positioning a write head on a canvas. Then, using the head to catalyze a local DNA strand-exchange reaction, they were able to selectively modify pixels on a canvas to write onto it.
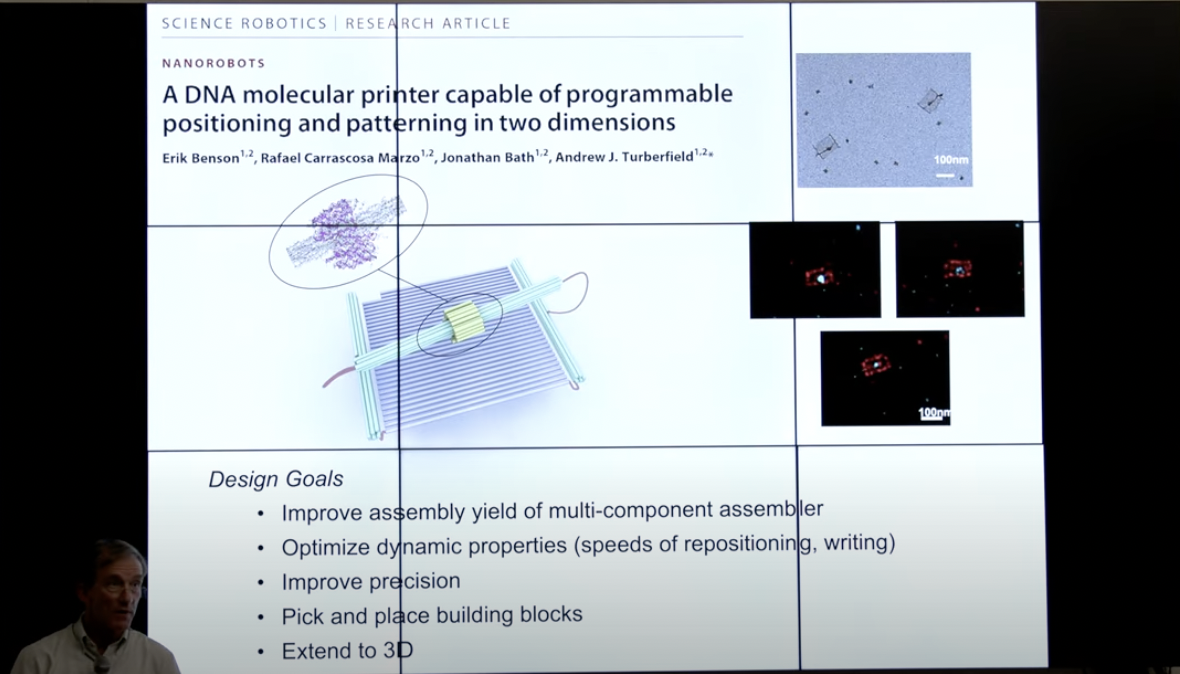
Although Tuberfield’s printer is only 2D, it points in the right direction for 3D printing. For such a structure to become a 3D printer, it would have to support precise movement along three axes to position the print head. Motion could be directed by external signals, such as exposure to light of different wavelengths, or by introducing other DNA strands that control individual components.
One of the materials that could be used in the printed products, are “spiroligomers,” robust and customizable building blocks similar to Lego bricks. Developed by chemist Chris Schafmeister, spiroligomers are synthetic molecules that are built on a rigid backbone from which further sidechains can be attached. Schafmeister has created a variety of spiroligomer shapes that can align and "click together" at specified binding sites, which is particularly useful when multiple binding locations are present.
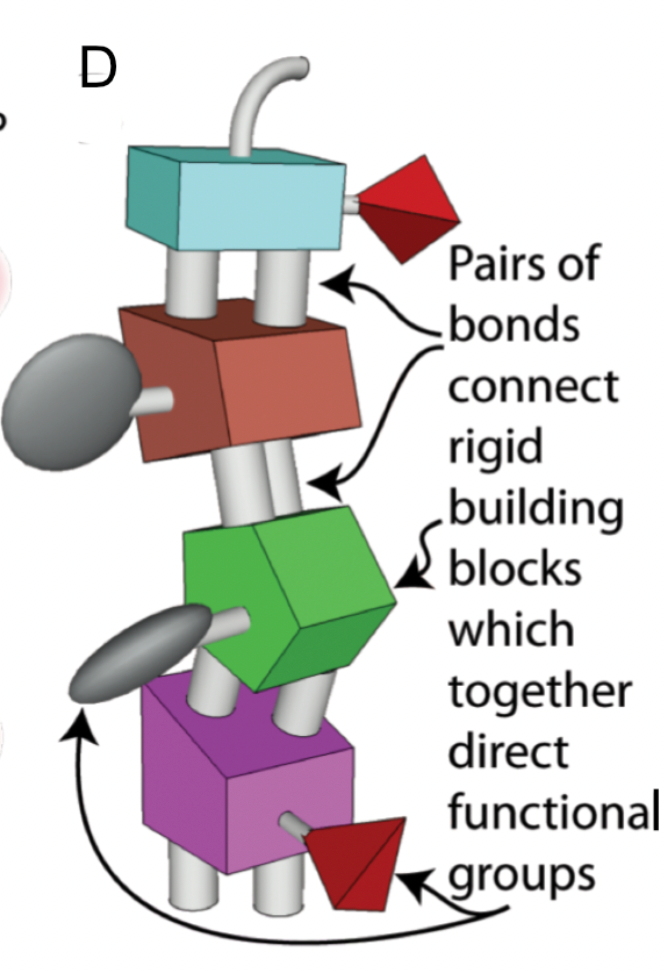
It may even be possible to design and fabricate spiroligomers using Schafmeister’s CANDO software. The software would allow designers to find spiroligomers which could bond with others as desired by finding matching reactive groups. A molecular printer would only need to bring the components into position, allowing the bond to form. 17 17 There are other design tools that are becoming available such as SAMSON, or MSEP. Expand Footnote Collapse Footnote
Using proteins for printing may also be possible, but is likely not as effective due to their complex structures and interaction types. Nevertheless, some researchers have focused on a subset of proteins with predictable shapes. For instance, David Baker’s lab recently demonstrated protein-protein binding with promising properties similar to DNA origami.
Are there other designs akin to the molecular 3D printer? A recent Foresight workshop explored a few alternative architectures, including the "molecular breadboard." This design also incorporates molecular building blocks, where a precise structure is built by utilizing selective bonding points on both the breadboard and the blocks. One general challenge with self-assembly methods is the need to encode the final structure in the specific interactions between blocks, which can be complex for larger structures. The molecular 3D printer, the molecular breadboard, and other emerging architectures aim to bypass this by merging self-assembly with precise positioning.
Despite these promising developments, there are still obstacles to overcome before we can assemble these components into a functional prototype. An earlier National Academy of Sciences (NAS) report expressed skepticism that merely modeling advanced molecular machines doesn’t provide a clear path to their construction. The report emphasized the need to focus on experimental steps and proof-of-concept studies. Such experiments and studies would require streamlined cooperation across diverse fields—physics, chemistry, and computer science—with potentially long time lags before results become clear.
Societal Roadblocks
The reason progress has stalled on molecular manufacturing isn’t due to technical problems alone—a series of social factors are adding to the difficulty. To understand why, it helps to look at the institutional history of nanotechnology.
In 2000, President Bill Clinton, inspired by the emerging possibilities of nanotechnology, launched the National Nanotechnology Initiative (NNI), which contributed billions of dollars to nanotechnology research. According to the NNI, the term nanotechnology encompasses everything from 1 to 100 nanometers. Despite early signals that the NNI would focus on research at the smaller end of the nanotechnology scale, it has mostly focused on advancing projects at the larger end which is far more tractable and the realm of near-term applications like nanomaterials. The smaller end of the scale, close to the size of an atom, is far more impactful, involving projects like molecular manufacturing. Unfortunately, working at the small end of the spectrum is very difficult, with the results being distant and expensive. The NNI’s definition encompasses a massive range of initiatives, with plausible, incremental, and established research crowding out ambitious projects. The initiative has undoubtedly contributed to many advances, such as the use of lipid nanoparticles to deliver mRNA vaccines. However, despite these serious successes, more ambitious molecular manufacturing goals have been left behind. This has led to much of the lower-hanging fruit being picked, with the transformational research left to rot on the upper branches. Generalist approaches that focus on molecular manufacturing should be prioritized over piecemeal proposals that focus on one particular application.
There is currently no existing organization with both the vision and funds required to tackle molecular manufacturing. Individual labs continue to apply for low-risk grants in tightly straightjacketed departments, with close to no incentives for collaboration across labs—let alone scientific fields—to generate the interdisciplinary proposals required for molecular manufacturing to succeed. Marblestone estimated that it would take around several research labs each five years and $50 million to develop a molecular printer, while Drexler estimated it would take 3-6 groups and $10-20 million per year for 3-5 years to develop. 18 18 See this conversation from 2016. Expand Footnote Collapse Footnote Without incentivizing collaborative efforts, molecular manufacturing will remain unrealized—even today when we have many of the building blocks in our hands.
New institutions are starting to emerge. Apart from long-standing nanotechnology organizations such as The Foresight Institute, 19 19 Founded in 1986 by Eric Drexler and Christine Peterson. Expand Footnote Collapse Footnote Adam Marblestone recently launched Convergent Research, a nonprofit supporting ambitious scientific projects that are neglected by legacy funders. 20 20 It starts from the observation that neither CERN nor the Hubble Space Telescope—both projects that are regarded as crucial public goods advancing science—were projects that individual academic labs, or private funders would have taken on. Expand Footnote Collapse Footnote To facilitate such much-needed innovations across disciplines, Convergent Research supports Focused Research Organizations (FROs) that are bigger than an academic lab, more coordinated than a consortium, and not directly profitable. Speculative Technologies is another recent example of a forward-thinking nonprofit that supports technical program managers who possess an ambitious scientific vision. Benjamin Reinhardt, head of the organization, helps them roadmap, set up, and run a coordinated research program across multiple organizations. 21 21 In 2021, Marblestone and Reinhardt joined a Foresight Institute seminar to jointly explore the Molecular 3D Printer as a concrete example of the kinds of ambitious goals they would like to see more of in the scientific ecosystem. Expand Footnote Collapse Footnote
Marblestone suggests that the funding problem around molecular manufacturing is a problem that most general-purpose technology encounters: for any product that already enjoys demand, there is likely a bespoke production method already in place. This means that basic research and development is extremely unprofitable before it is sufficiently advanced to compete with specialized production methods. As a result, innovators face a high temptation to focus on immediate end results, rather than the underlying process that has broad applications. Sometimes, the urgency of the moment calls for the shortest path rather than fundamental research and general tool development. For instance, current mRNA vaccines are delivered in lipid nanoparticles—bits of RNA wrapped in fat. It’s possible that advanced nanotechnology could produce more potent and flexible vaccines, but we don’t have time for such research during a pandemic. 22 22 See this section of Marblestone’s analysis. Expand Footnote Collapse Footnote
The debate between Eric Drexler and Richard Smalley in the early 2000s encapsulates the tension between piecemeal and generalist approaches. Smalley won a Nobel Prize in chemistry for his discovery of “buckyballs,” a carbon molecule with remarkable properties. Smalley supported research on incremental nanomaterial improvements and was skeptical of molecular manufacturing.
Arguably, Smalley was attacking a strawman by criticizing molecular manufacturing as if it called for specific capabilities. For instance, one of his main objections was that mechanosynthesis would be “fat-fingered,” preventing a mechanically guided assembler from releasing an atom at a precise time and place. Drexler and colleagues laid out plenty of paths to molecular manufacturing that would avoid such a problem.
Another idea that did lasting damage to molecular manufacturing’s reputation was the idea of rogue nanobots. In a 2000 Wired article, Sun Microsystems CEO Bill Joy posited that self-replicating nanobots could constitute a serious threat to human existence by escaping human control and converting all available matter into “gray goo.” 23 23 There is a strong parallel here between “gray goo” and the “paperclip maximizer” of AI safety. Both are about technology escaping human control and using all available matter for some inscrutable purpose. Expand Footnote Collapse Footnote There are many good reasons to believe that the gray goo scenario has been overstated. For instance, molecular manufacturing could come with restrictions so that their products are incapable of self-replication or self-sufficiency. 24 24 Such simplistic sounding solutions are possible if considered in isolation from possible AI alignment issues that might emerge from the “engines of design” side of the equation. Expand Footnote Collapse Footnote
Although the risks from gray goo are likely minimal for the foreseeable future, there are other risks worth worrying about. Molecular manufacturing would basically make everything easier and cheaper to produce—centrifuges for enriching uranium could become far more accessible. The problems related to molecular manufacturing are less about gray goo and more about a lower barrier for malicious actors, a perennial problem in technology development. 25 25 Chris Phoenix argues that the potential for dangerous nanoscale applications may be best considered in the context of military competition and arms control. Also, see Nick Bostrom’s description of “easy nukes” in “The Vulnerable World Hypothesis” Expand Footnote Collapse Footnote On the other hand, the potential of molecular manufacturing to create material abundance could also reduce pressures that drive global conflicts.
The Future of Molecular Manufacturing
In 1993, Ralph Merkle asked conference attendees how long they thought it would take to develop molecular nanotechnology; the answers ranged from 2010 to 2040. In a 2014 interview, Drexler notes that if there was agreement on a well-structured research program and sufficient funding, we could potentially get there within 10 years. Even though we are not there yet, three key ingredients are now in place for realizing molecular manufacturing within our lifetimes. First, tools and components are now available which could be combined into a molecular 3D printer prototype. Second, software tools that simplify the complex design process are becoming operational. Third, there is renewed appetite from nonprofit institutions and a few ambitious individual actors to do the actual work of assembling a prototype.
It’s unfortunate that much of the effort to reignite progress in nanotechnology is coming from nonprofits and individual actors, when such a project is arguably the mandate of the government. Lack of government support in fundamental research invites questions about stagnation that have become so popular recently. One hope is that building functional prototypes might lead to greater investment from institutions both private and public.
Amazing things can happen when researchers are given the opportunity to focus on projects that are not of immediate utility—Bell labs, or Xerox PARC, stand as particularly productive examples of fundamental research. 26 26 This problem has been long acknowledged, for instance see Abraham Flexner in "The Usefulness of Useless Knowledge”, published in 1939. Expand Footnote Collapse Footnote Bell Labs contributed to incredible advances across multiple industries, including the transistor, the solar cell, and communication satellites. Xerox PARC developed many components that would be combined in modern computers and networks. Molecular manufacturing still requires substantial research and development before it becomes operational, but it would be one humanity’s greatest achievements, transforming almost every industry imaginable.
Physical engineering needs to stage a comeback. If we desire the world to be different, it may mean prioritizing atoms over bits for a while—or at least putting the bits to work on the challenge. Molecular manufacturing may be ambitious, but it was expected to arrive much sooner. Let's focus on the tiny details of the real world we live in, so that we can manufacture a better one tomorrow.